Study on glucose metabolism by stable isotope labeling
Ron Orlando
Center for complex carbohydrate research, Department of Biochemistry and molecular biology, Department of chemistry, University of Georgia, Athens, GA 30602 USA
"My lab has used a series of CIL products, and their products perform well in proteomics and glycomics. CIL's salesmen are very helpful and knowledgeable. They have helped us with the experimental design many times. Without their help, we could not have completed this research work. " --– Ron Orlando
Glycosylation is one of the most common protein expression modifications in eukaryotic system. It is estimated that 60-90% of mammalian proteins are glycosylated. In fact, all membrane and secretory proteins are also glycosylated. Glycoproteins usually play key roles in physiological metabolism, such as cell recognition, signal transduction, inflammation and carcinogenesis. In view of the important physiological role of protein glycosylation, many research teams are committed to the structural identification of specific glycans, the expression of protein glycans, and the detailed study of how these structures change, such as cell differentiation or with the development of tumor cells. All these efforts have led to a new field of research glycomics.
Mass spectrometry (MS) has been developed as a method for the analysis of complex mixtures, which is popular for its rapidity, sensitivity and accuracy. Glycomics research usually involves mass spectrometry (MS) analysis of the complete decomposition products of glycans. Although the qualitative analysis of glycan is an effective method, the quantitative analysis of glycan by MS is controversial. For example, matrix effect, due to the ion inhibition of other compounds, can affect the analysis results of the specified substances without changing the substrate concentration. Other factors that interfere with the results of mass spectrometry analysis include the detection limits of different instruments, the errors of instruments, and the loss in the process of sample processing. The successful application of different relative quantitative methods depends on how to solve the error of the original data.
The use of isotope labeling technology can make up for these quantitative defects, so it has been widely used in various "omics", especially in the field of proteomics. Stable isotope labeled amino acids are a good example in cell culture (SILAC). This method perfectly combines isotope labeled amino acids with proteins to study proteomics by mass spectrometry. In the SILAC experiment, two kinds of cells were grown in the medium, one with "light amino acids" (natural abundance), the other with heavy amino acids (isotope labeling) (for example, unlabeled and L-Lysine · 2HCl (13C6, 99%) (CIL catalog number clm-2247) and L-arginine · HCl (13C6, 99%) (CIL catalog number clm2265)). Isotope labeled amino acids replace the natural amino acids used in cell culture and bind to all new and mature proteins. In the process of cell division, each specific amino acid is replaced by the corresponding isotope labeled amino acid. The advantage of this method is that the two cell types are mixed immediately after lysis, so the experimental conditions of digestion, purification and separation of proteins from the two cell types are exactly the same in the same sample processing. For this reason, SILAC is generally regarded as the gold standard for proteomic quantification.
In this paper, an in vivo labeling method for glycomics is described, which is similar to the application of SILAC in proteomics. This glycomics method is called idawg - (glutamine to amino sugar isotope detection) - based on the fact that the glutamine side chain is the only nitrogen donor in the synthesis of amino sugar nucleotides (Fig. 1). Therefore, when L-glutamine (amide-15n, 98%) was introduced into the medium without glutamine, all amino sugars including acetylglucosamine, acetylgalactosamine and sialic acid labeled by N15 could be produced. This resulted in an increase of + 1da in the mass of N and O-containing glycans, glycolipids and extracellular matrix polysaccharides. This method was verified by the experiment of releasing N-containing glycan from protein of mouse embryonic stem cells cultured in unlabeled and N15 labeled glutamine environment. The success of these experiments enables us to predict that idawg technology will be helpful to the study of various comparative glycomics in cell culture in the future.
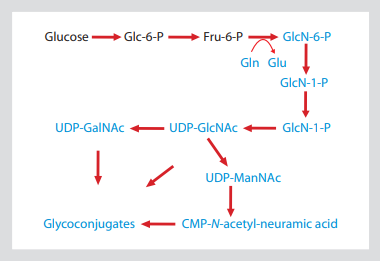
Fig. 1: hexosamine biosynthesis pathway shows that the side chain of glutamine is the ammonia in the process of glyconucleotide production
This allows 15N isotope labeling to be introduced into all amino sugars, including amino sugars
GlcNAc, GalNAc and sialic acid. The substance containing 15N is shown in blue.
conclusion
The first and rate limiting step of hexosamine biosynthesis pathway (Fig. 1) is the transformation of glycolysis intermediate (fructose-6-phosphate to glucosamine-6-phosphate). The amino nitrogen introduced in this step is only supplied by the side chain amide of Gln (glutamine), which is converted into Glu (glucose). Glucosamine-6-phosphate is the precursor of UDP GlcNAc, which in turn leads to other major sugar containing nucleotide amino sugars udpgalnac and CMP sialic acid. Therefore, all the molecules containing GlcNAc -, GalNAc - and sialic acid are isotope labeled targets by adding amide-15n-gln to cell culture medium.
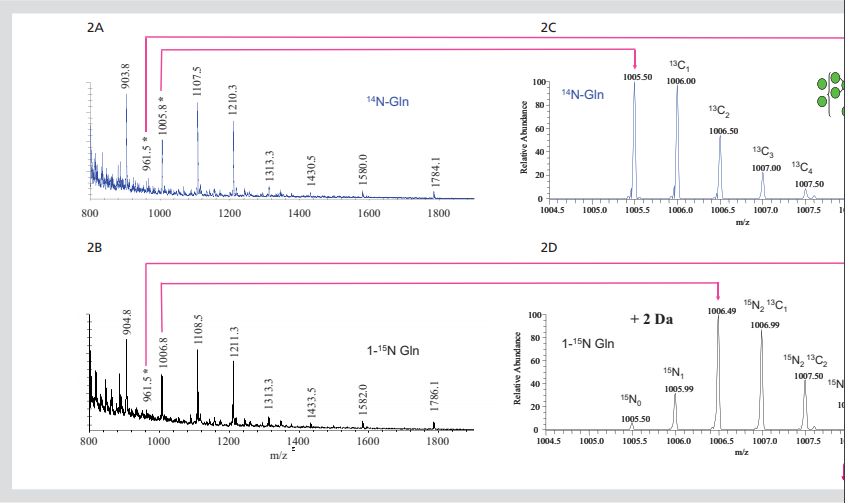
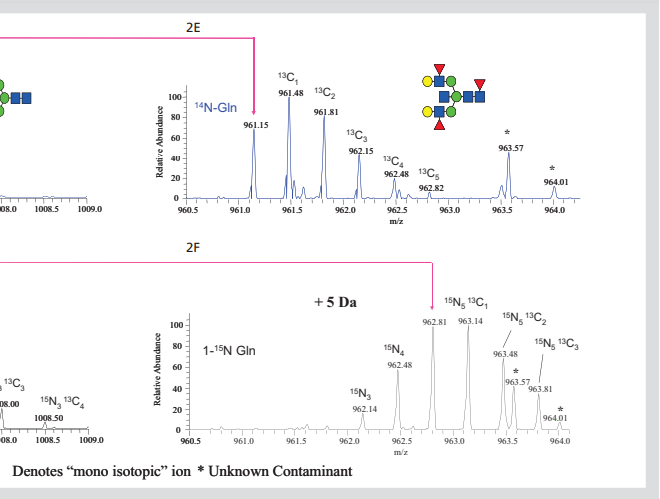
Figure 2. Isotope labeled N-linked glycans. All ft-ms spectra of 850-2000m / Z dimethyl n-chain
Glycan (2a) or amide-15n-gln (2b) released by cells growing in labeled Gln. Extended region of spectrum
The predicted 2 Da mass shift of 15N into the two core GlcNAc residues of glcnac2man7 (2c,
2D) and 5-da increased, corresponding to the incorporation of 15N into glcnac5man3gal2fuc3 glycan (2e, 2f).
Preliminary experiments were conducted to evaluate the possibility of implanting stable isotopes into glycans of cultured cells using metabolic labeling method. In these experiments, R1 mouse embryonic stem cells (mESCs) were cultured under standard conditions. Cell culture media usually do not contain glutamine because this amino acid is decomposed into glutamic acid and ammonia in aqueous solution. This method simplifies the idawg labeling method because Gln (glutamine) medium does not need to be specially consumed. The medium was supplemented with 15N labeled - Gln or unlabeled Gln at a standard concentration (2 mm). This direct substitution is the only idawg needed to change the normal cell culture procedure. In this case, mouse embryonic stem cells were cultured with labeled or unlabeled Gln for three days, and then n - and O-linked glycans were isolated from the protein for mass spectrometry analysis. The incorporation of 15N into the N-linked glycans of MESC was investigated by comparing ft-ms (ultra high resolution spectroscopy) spectra of hypermethylated N-linked glycans released by cells grown in unlabeled Gln or L-glutamine (amide-15n, 98%) (FIGS. 2a and 2b). The abundant ions observed in these spectra correspond to single or double charged high mannan (glcnac2man5-9). The comparison of the two spectra showed that the double charged ions increased by 1 m / Z unit and the single charged ions increased by 2 m / Z unit. This is expected if 15N has been incorporated into the two core GlcNAc in these glycans, and this is the only nitrogen in high mannose glycans.
This change in the mass of glycan can be clearly seen by careful observation of the double charged molecular ion (M + 2Na) 2 +, which is produced by glcnac2man7 glycan and appears in 14N and amide-15n glycans in 1005.5 and 1006.5 M / Z units, respectively (Fig. 2C and 2D). In these spectra, the most responsive ions appear at the single isotope mass calculated by 14N or two 15N, which indicates that most of these glycans have 15N binding to two possible sites. There was an ion corresponding to the lack of labeling, that is, a combination of zero and a 15N; however, the most responsive one was completely labeled.
A large number of N-linked glycans were similarly increased in mass by inserting a 15N into each GlcNAc terminal group. This can be observed by extending the spectra shown in FIGS. 2a and 2b so that only regions from 960.5 to 964.1 can be observed (FIGS. 2e and 2F, for glycans released by cells grown in 14N and amide 15N Gln media, respectively). This region shows molecular ion, (M + 3Na) 3 +, which is used to form complex glycans of glcnac5man3gal2fuc3. As predicted, when the glycan was obtained from cells grown in 15N Gln, its single isotope ion increased by + 5da. In addition, the fully labeled glycan is the main form of existence, but the ions can be observed from the incomplete labeled glycan.
By calculating the different isotopes of 15N spectral intensity, it is shown that the polysaccharide contains 98% 15N, which is exactly equal to the amount of 15N in glutamine used in this experiment. Similar calculations of isotopic distributions of other glycans show that their 15N incorporation ranges from 96% to 98%. These examples indicate that 15N in L-glutamine (amide-15n, 98%) is widely incorporated into the GlcNAc end group of N-linked glycans, suggesting that this may be an effective way to introduce stable isotopes into glycans.
discuss
Metabolic markers of glycans provide many new opportunities for evaluating the kinetics of glycan conversion during any cell behavior induced or maintained in culture. For example, labeling cells with L-glutamine (amide-15n, 98%) and then replacing the medium supplement with light Gln can determine the half-life of any aminoglycan. Previously, glycan transformation studies need to combine radiomonosaccharide and extensive subsequent fractionation to determine specific changes in glycan expression. Generally speaking, these radiotracer techniques can be very sensitive to detect polysaccharides, but they have low resolution in tracking the biosynthesis related substances of individual glycan structures or subgroups. The stable isotope incorporation method reported in this paper combines the advantages of high resolution mass spectrometry with the biological necessity of understanding the kinetics of glycan transformation. Therefore, idawg presents an effective quantitative method to explore the biological role of glycan, glycoprotein and glycolipid in cell culture system.
thank
This study was funded by the National Institutes of health / NCRR funded biomedical glycomics comprehensive technology resources (p41 rr018502) and the National Institutes of health / NCRR funded comprehensive sugar technology research resources (p41 rr005351).
SILAC related products
|
No.
|
Product Name
|
NO.
|
Product Name
|
|
CLM-2265
|
L-Arginine·HCl (13C6, 99%)
|
DLM-4212
|
L-LEUCINE (ISOPROPYL-D7, 98%)
|
|
NLM-557
|
L-Glutamine (amide-15N, 98%)
|
CLM-653
|
L-LYSINE:2HCL (1-13C, 99%)
|
|
CLM-2247
|
L-Lysine·2HCl (13C6, 99%)
|
CLM-653
|
L-LYSINE:2HCL (1-13C, 99%) MICROBIOLOGICAL/PYROGEN TESTED
|
|
DLM-6038
|
L-ARGININE:HCL (4,4,5,5-D4, 94%)
|
CNLM-7821
|
L-LYSINE:2HCL (1-13C, 99%; ALPHA-15N, 98%)
|
|
NLM-395
|
L-ARGININE:HCL MICROBIOLOGICAL(15N2,98%+)
|
CDNLM-6810
|
L-LYSINE:2HCL (13C6, 97-99%; D9, 97-99%; 15N2, 97-99%)
|
|
CLM-2051
|
L-ARGININE:HCL (1,2-13C2, 99%)
|
CNLM-291
|
L-LYSINE:2HCL (13C6, 99%; 15N2, 99%)
|
|
CLM-1268
|
L-ARGININE:HCL (1-13C, 99%)
|
NLM-1554
|
L-LYSINE:2HCL (15N2, 98%+)
|
|
CNLM-7819
|
L-ARGININE:HCL(1-13C,99%;ALPHA-15N,98%)
|
DLM-2641
|
L-LYSINE:2HCL (3,3,4,4,5,5,6,6-D8, 98%)
|
|
CDNLM-6801
|
L-ARGININE:HCL (13C6, 97-99%; D7, 97-99%; 15N4, 97-99%)
|
DLM-2640
|
L-LYSINE:2HCL (4,4,5,5-D4, 96-98%)
|
|
CNLM-539
|
L-ARGININE:HCL (13C6, 99%; 15N4, 99%)
|
DLM-2640
|
L-LYSINE:2HCL (4,4,5,5-D4, 96-98%)MICROBIOLOGICAL/PYROGEN TESTED
|
|
NLM-396
|
L-ARGININE:HCL (15N4, 98%)
|
CLM-632
|
L-LYSINE:2HCL (6-13C, 99%)
|
|
NLM-1267
|
L-ARGININE:HCL (ALPHA-15N, 98%+)
|
CNLM-3454
|
L-LYSINE:2HCL (6-13C, 99%; EPSILON-15N, 98%)
|
|
DLM-541
|
L-ARGININE:HCL (D7, 98%)
|
NLM-143
|
L-LYSINE:2HCL (ALPHA-15N, 98%)
|
|
DNLM-7543
|
L-ARGININE:HCL (D7, 98%; 15N4, 98%)
|
NLM-143
|
L-LYSINE:2HCL (ALPHA-15N, 98%) MICROBIOLOGICAL/PYROGEN TESTED
|
|
CLM-2070
|
L-ARGININE:HCL (GUANIDO-13C, 99%)
|
DLM-570
|
L-LYSINE:2HCL (D9, 98%)
|
|
NLM-395
|
L-ARGININE:HCL (GUANIDO-15N2, 98%+)
|
DNLM-7545
|
L-LYSINE:2HCL (D9, 98%; 15N2, 98%)
|
|
ULM-8347
|
L-ARGININE:HCL UNLABELED
|
NLM-631
|
L-LYSINE:2HCL (EPSILON-15N, 98%+)
|
|
CLM-2262
|
L-LEUCINE (13C6, 99%)
|
ULM-8766
|
L-LYSINE:2HCL UNLABELED
|
|
CLM-2262
|
L-LEUCINE (13C6, 99%) MICROBIOLOGICAL/PYROGEN TESTED
|
|
|
|
CNLM-281
|
L-LEUCINE (13C6, 99%; 15N, 99%)
|
|
|
1. Varki, A. 1993. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology, 3(2), 97-130.
2. Krueger, K.E.; Srivastava, S. 2006. Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol Cell Proteomics, 5(10), 1799-1810.
3. Apweiler, R.; Hermjakob, H.; Sharon, N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta, 1473(1), 4-8.
4. Drickamer, K.; Taylor, M.E. 1998. Evolving views of protein glycosylation. Trends Biochem Sci, 23(9), 321-324.
5. Dwek, R.A. 1995. Glycobiology: more functions for oligosaccharides. Science, 269(5228), 1234-1235.
6. Lis, H.; Sharon, N. 1993. Protein glycosylation. Structural and functional aspects. Eur J Biochem, 218(1), 1-27.
7. Haltiwanger, R.S. 2002. Regulation of signal transduction pathways in development by glycosylation. Curr Opin Struct Biol, 12(5), 593-598.
8. Lowe, J.B. 2003. Glycan-dependent leukocyte adhesion and recruitment in inflammation. Curr Opin Cell Biol, 15(5), 531-538.
9. Fukuda, M. 1996. Possible roles of tumor-associated carbohydrate antigens. Cancer Res, 56(10), 2237-2244.
10. Hakomori, S. 1989. Aberrant glycosylation in tumors and tumor-associated carbohydrate antigens. Adv Cancer Res, 52, 257-331.
11. Lowe, J.B.; Marth, J.D. 2003. A genetic approach to mammalian glycan function. Annu Rev Biochem, 72, 643-691.
12. Muramatsu, T. 1993. Carbohydrate signals in metastasis and prognosis of human carcinomas. Glycobiology, 3(4), 291-296.
13. Olden, K. 1993. Adhesion molecules and inhibitors of glycosylation in cancer. Semin Cancer Biol, 4(5), 269-276.
14. Lubner, G.C. 2003. Glycomics: an innovative branch of science. Boll Chim Farm, 142(2), 50.
15. Zaia, J. 2004. Mass spectrometry of oligosaccharides. Mass Spectrom Rev, 23(3), 161-227.
16. North, S.J.; Koles, K.; Hembd, C.; et al. 2006. Glycomic studies of Drosophila melanogaster embryos. Glycoconj J, 23(5), 345-354.
17. Jang-Lee, J.; North, S.J.; Sutton-Smith, et al. 2006. Glycomic profiling of cells and tissues by mass spectrometry: fingerprinting and sequencing methodologies. Methods Enzymol, 415, 59-86.
18. Ong, S.E.; Blagoev, B.; Kratchmarova, I.; et al. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics, 1(5), 376-386.
19. Ong, S.E.; Foster, L.J.; Mann, M. 2003. Mass spectrometric-based approaches in quantitative proteomics. Methods, 29(2), 124-130.
20. McClain, D.A. 2002. Hexosamines as mediators of nutrient sensing and regulation in diabetes. J Diabetes Complications, 16(1), 72-80.
21. Yki-Jarvinen, H.; Vogt, C.; Iozzo, P.; et al. 1997. UDP-N-acetylglucosamine transferase and glutamine: fructose 6-phosphate amidotransferase activities in insulin-sensitive tissues. Diabetologia, 40(1), 76-81.
22. Kean, E.L.; Munster-Kuhnel, A.K.; Gerardy-Schahn, R. 2004. CMP-sialic acid synthetase of the nucleus. Biochim Biophys Acta, 1673(1-2), 56-65.
23. Thoden, J.B.; Wohlers, T.M.; Fridovich-Keil, J.L.; et al. 2001. Human UDPgalactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J Biol Chem, 276(18), 15131-15136.




